摘要
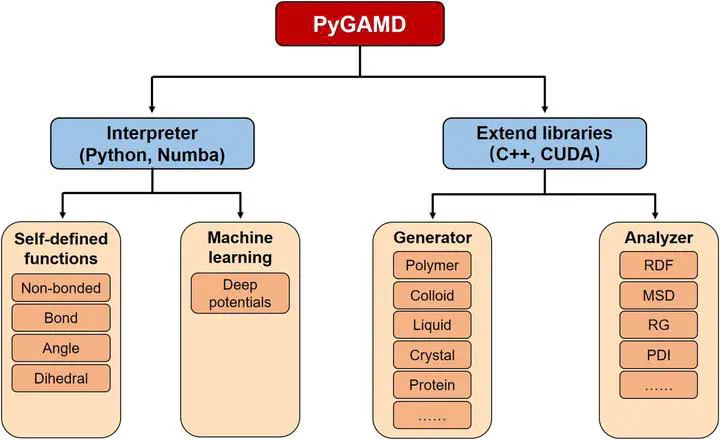
PyGAMD (Python GPU-accelerated molecular dynamics software) is a molecular simulation platform developed from scratch. It is designed for soft matter, especially for polymer by integrating coarse-grained/multi-scale models, methods, and force fields. It essentially includes an interpreter of molecular dynamics (MD) which supports secondary programming so that users can write their own functions by themselves, such as analytical potential forms for nonbonded, bond, angle, and dihedral interactions in an easy way, greatly extending the flexibility of MD simulations. The interpreter is written by pure Python language, making it easy to be modified and further developed. Some built-in libraries written by other languages that have been compiled for Python are added into PyGAMD to extend it’s features, including configuration initialization, property analysis, etc. Machine learning force fields that are trained by DeePMD-kit are supported by PyGAMD for conveniently implementing multi-scale modeling and simulations. By designing an advanced framework of software, graphics processing unit-acceleration achieved by the Numba library of Python and compute unified device architecture reaches a high computing efficiency.